MALDI-MSI in Pharmaceutical R&D
MALDI-MSI is becoming more widely adopted in pharmaceutical research and development
A major limitation to the quantification of drug candidates in blood and tissues with liquid chromatography coupled to mass spectrometry (LC-MS), and other widely used analytical techniques is the lack of spatial information. Often, the drug concentration measured in plasma is not representative of its concentration within various tissues, and this has significant implications for efficacy or toxicity assessment. Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) is gaining momentum in pharmaceutical research and development. This label-free technology enables quantitative spatial analysis of drug candidates in tissues. Direct profiling of proteins in cryosectioned tissue was achieved in 1999, and it has since been applied to organs and whole-body animal sections. With further refinement, and the development of standardized approaches and reporting guidelines, MALDI-MSI may become an essential technology for drug development.
Working with Cryosections
Dr. Jonathan Stauber (founder) and Dr. Erin Seeley (principal scientist) at ImaBiotech, a contract research organization with facilities in Boston, MA and France, use MALDI imaging for drug efficacy and toxicity evaluations. “We use MALDI imaging to determine the level of drug exposure to specific cells such as tumor cells. In certain cases, we have been able to demonstrate a drug reaching a tumor biopsy has a limited exposure to the tumor cell and limited efficacy. We go further than classical methods to evaluate the drug efficacy,” explain Stauber and Seeley.
Unlike traditional LC-MS techniques that require sample homogenization and analyte extraction, MALDI-MSI analyses require frozen tissue sections fixed to a slide. According to Stauber and Seeley, “in a MALDI imaging experiment, thin sections of tissue are collected, similar to those used for pathology, onto special slides that are compatible with a mass spectrometer.” Part of sample preparation involves coating the tissue sample with a matrix: “typically a small organic, aromatic compound that can absorb the energy from the laser and transfer it to the molecules in the tissue allowing them to be desorbed and ionized.” The laser is rastered—forming a grid over the sample—and generates pixels of data, “just like the pixels in a picture or television, but with hundreds to thousands of analytes measured simultaneously from each pixel. Each analyte can then be displayed as a function of its position and relative intensity on the tissue section,” they explain. Using MALDI-MSI, the target analyte’s spatial location is preserved, providing valuable data pertaining to toxicology. “An example of where this is advantageous is if an analyte of interest is highly concentrated in only one area of the tissue (e.g. the bile ducts of the liver), but is absent from the rest of the tissue. The high concentration in one area could lead to toxicological effects. In an LC-MS approach, the average concentration in the liver would be low, and would make it difficult to determine why adverse effects were occurring,” explains Stauber.
Quantitative Capabilities
MALDI-MSI can be used to determine the relative concentration of an analyte in tissue samples. Pharmaceutical research often necessitates absolute quantification, which may be achieved with quantitative MSI (qMSI). qMSI requires a calibration curve and an internal standard. “The internal standard is used to normalize the spectral intensity of each pixel of the image and can help to correct for any differences in ionization that occur in different tissues (e.g. tumor, stroma, normal, etc.),” says Seeley. Often, a stable-isotope labeled version of the analyte is added to the matrix solution, and the analyte’s ion intensity is normalized to the standard.
There are two common approaches to creating a calibration curve; one involves creating a spotted dilution series of standards on the surface of a control sample. Alternatively, mimetic tissue models may be created by homogenizing control samples with a range of drug concentrations, and freezing them in a cylindrical mold. This approach is thought to more accurately represent analyte extraction and ion suppression in dosed tissue. Regardless of the approach, it is essential that the results correlate with LC-MS, currently the gold standard for quantification.
Managing Enormous Amounts of Data
As with other MS methods, MALDI-MSI generates enormous amounts of data, with thousands of spectra generated for each pixel. Software enables the spectra to be compiled into a single image file for visualization. “Depending on the goals of the study, there are different ways the data are analyzed,” explains Seeley. “For a biomarker discovery study, peaks are interrogated from the average spectra to determine those that show spatial distribution/concentration that is relevant to the biology being studied (e.g. tumor area vs. normal area, treated sample vs. untreated sample, etc.). Alternatively, for a drug or metabolite quantification study, the data from the targeted molecules of interest is extracted and a calibration curve is applied to determine the amount of the molecule in either g/g of tissue or μM concentration in tissue. Quantification can be carried out on specific regions to compare with histology or to correlate with other tissue markers,” says Seeley.
A Couple of Caveats
“The biggest challenge associated with MALDI imaging is sensitivity for the target molecule(s). Since the analysis is done in situ with no chromatographic separation of analytes, ion suppression effects due to the considerable dynamic range of analytes in the tissue can be detrimental to the detection of the desired molecule(s),” says Stauber.
Incorporating an internal standard can correct for ion suppression, and adding a deuterated analog of the molecule of interest is considered the best method. Ensuring a homogenous deposition of matrix is also essential for successful MALDI imaging. Air-sprayers are the most widely used matrix application devices, and achieve a homogenously coated tissue sample. Spraying also produces smaller droplets compared to other techniques, resulting in smaller matrix crystals that provide higher spatial resolution.
Another challenge is the inability to concentrate or amplify the signal with MALDI imaging. “Tissue sections are typically 10 μm in thickness and the ablated area for each pixel is generally 30-100 μm in diameter. This means that there may be very few molecules of the target analyte within each sampled area. If the analyte concentration is too low in an ablated area, it will not be detected,” explains Seeley.
While MALDI-MSI is not without challenges, the technique has clear benefits for pharmaceutical research and development. “One of the major advantages of MALDI imaging over other methods is that it is a label free technology, and no analyte specific reagents are needed,” says Stauber. He adds: “MALDI imaging can also be relatively high throughput in that several different tissue sections can be placed on a single MALDI-compatible slide and imaged in a single experiment.”
Emerging Applications
MALDI imaging shows potential as an approach to examining the off-target effects of drugs. Part of the process of drug development is ensuring that lead compounds have high affinity for targets, and that they do not interact with other non-targets throughout the body. Off-target drug interactions have been identified using Bead-GPS Drug-Protein Screening, by which compound libraries are screened against protein libraries. Using this technique, a photocleavable mass-tag encoded protein-bead library is incubated with a compound library, and MALDI-MSI is used to decode beads with photocleaved mass-tags and identify compounds bound to the bead using molecular weight.
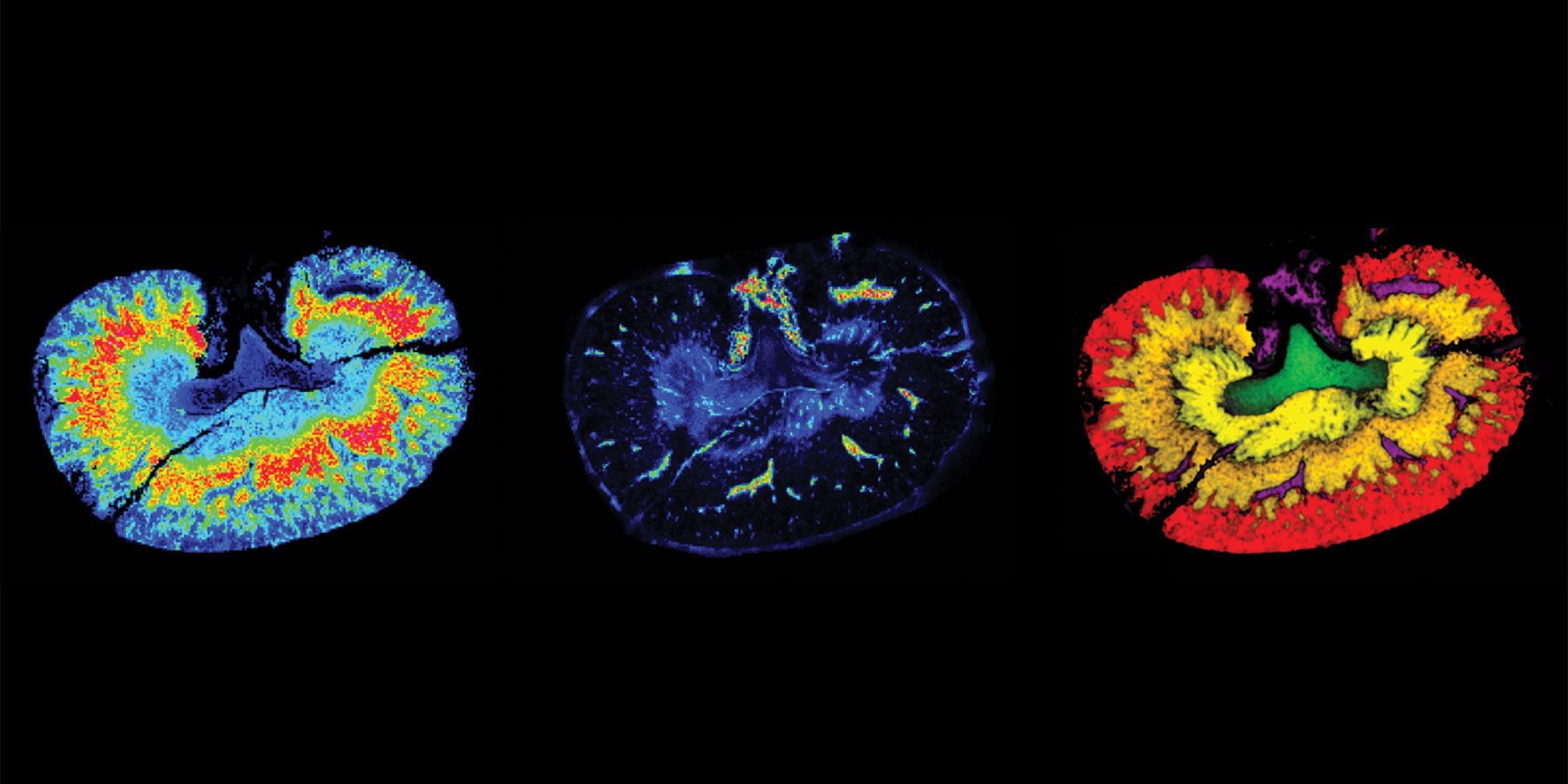
Combined with other analytical techniques, MALDI-MSI also shows potential for solving toxicological problems in drug development. The technique has been shown to effectively detect, map, and identify molecular differences between drug treated and control tissue sections (in this case, kidney tissue), making it a valuable tool for the identification and distribution of compounds in preclinical toxicology studies.
MALDI-MSI capabilities extend beyond quantitative spatial analysis of target compounds to include target engagement and toxicity, among others. As instrumentation and methods continue to advance, it becomes more widely adopted in pharmaceutical research and development.

