All Methods Still Enjoy Substantial Success
Instead of seeking to provide a comprehensive overview of the various optical and non-optical methods, we instead discuss the work of three leading research groups.
Several research centers have sprung up to work on cutting-edge research. The Howard Hughes Medical Institute Janelia Research Campus in Ashburn, Virginia, is a leading global center for basic research and product development for all aspects of live-cell imaging (LCI). The late Mats Gustafsson led a team of scientists who invented aberration-corrected multi-focus microscopy, which allows investigators to image live cells simultaneously. Another Gustafsson brainchild, a live-cell multicolor structured illumination microscope, uses interference patterns generated when grated light illuminates fine biological structures to deliver resolution at 110 nanometers. What sets this system apart is its maximal speed at six optical sections/microns/ second, making it perfectly suited for live-cell applications.
We asked Teng-Leong Chew, PhD, director of the Advanced Imaging Center at Janelia, what characteristics the ideal LCI microscope might possess. “The instrument must be fast enough to capture the event under study, and the laser illumination should be gentle.”
Speed is a relative concept in biology. Cell-imaging systems generally operate from about ten frames per second (fps) to about 20 frames per minute. The faster time frames enable visualization of fast events like microtubule- based transport and neuronal firing. “If you’re only looking at a cell crawling it can be a bit slower, and for cell division slower still, depending on which event within that process you’re observing.”
Researchers want their camera to be as sensitive as possible, the imaging probe to be as bright as possible, and the signal-to-noise (S/N) ratio to be as high as possible. This means illumination light needs to be as gentle on cells as possible, given the experimental goals. The higher the energy load on cells, the shorter the observation period.
Chew calls lattice light sheet microscopy (LLSM), invented by Janelia applied physicist and Nobel Prize winner Eric Betzig, “the most important live-cell instrument introduced in 20 years. It delivers very gentle illumination power that permits high-speed, 3-D, multicolor, and long-term imaging of live cells.”
Living cells can be visualized in “volumes,” which are reassembled and rendered as image slices or sections similar to those in medical imaging. With conventional microscopy, cells die after 20–30 volumes. With LLSM, cells can withstand imaging up to 15 to 20 thousand volumes, several orders of magnitude higher than through conventional techniques. “And the signal-to-noise [ratio] is so good you get details you didn’t see before,” Chew tells Lab Manager. “Conventional live-cell imaging systems are fast at sectioning cells, but LLSM is much faster, and gentler as well,” he says.
LCI began in earnest with widefield microscopy, in which light illuminates an entire specimen. Think about this: The camera collects all the reflected light, which does not produce great images. “If your cells are thin, then widefield microscopy doesn’t pose much of a problem,” Chew explains, “but if they’re thicker, you see a lot of light that is out of focus. The camera collects everything. It’s a gentle method but details are poor, especially in the z-direction.”
Laser scanning or spinning disk confocal microscopy does a slightly better job by using pinholes to reject outof- focus light. Images are sharper than with widefield, but the pinhole rejects a lot of otherwise informative photons, so the technique is extremely wasteful in terms of the cells’ light exposure.
“To slice cells optically with spinning disk you must move the microscope stage up or down, illuminate, reject most of the incoming photons, capture the image, and move the stage again,” Chew says. “This happens very fast, but cells sitting on the microscope stage are being exposed to the repeated high-intensity laser, and most of the photons it returns are rejected. Eventually the cell dies.”
Cell Health by Reducing Exposure to Laser Light
LLSM, a notable improvement, interrogates the sample as a very thin sheet, illuminating just a very thin slice. The rest of the cell remains dark. No pinhole is required because all the emitted light comes from the same thin focal plane, and the camera collects every photon that returns to the microscope. “The cell remains healthy because [you] don’t see as many photons as in other methods, and since the illuminated layer is so thin you get very good S/N [ratio],” Chew explains.
An emerging work-around involves the use of two-photon excitation, which allows the use of two lower-energy (longer-wavelength) photons whose effect is additive. “Two-photon systems are suitable to live-cell imaging and are less damaging to cells or tissues,” James Lopez, PhD, national sales support specialist at Olympus (Center Valley, PA), explains. “You can therefore image for longer with less photon damage in the longer wavelengths. The technology to move out to longer and longer wavelengths has only really matured during the past two years. Some companies are offering two-photon systems that go out to 1,300 nanometer excitation, and they’re imaging much faster, which is easier on the cells.”
Like single-photon techniques, two-photon excitation works by exciting the pi electrons inside the dye, which upon relaxation emit at a higher wavelength. The difference is that two-photon techniques squeeze photons into a very small excitation volume. “The fluorescence is the same, but we collect light differently with two-photon [excitation],” Lopez says. Single-photon excitation uses a pinhole to exclude out-of-focus light, but with two photons the excitation spot itself is volume- limited, so every photon collected is in focus.
All other things being equal, resolution improves at shorter wavelengths, and that is a plus for high-speed dynamic imaging. But the situation is not quite that straightforward. “It depends on how much power the system needs to generate an image for that application,” Lopez tells Lab Manager. “If you’re scanning super, super fast and your fluorophore doesn’t emit enough light during that scanning period, you’re not getting an image.” industry insights: life science
Optimized for Speed
Lopez has worked on developing scanning systems optimized for speed. One of those is a resonance device operating around 438 fps, within range of the speed of a resonance scanner. “That’s one way to go fast,” Lopez says: spinning disk technology, CSU, microlens-based spinning disk systems that, instead of scanning the beam left to right across the sample, essentially have thousands of microbeams that are scanned across the sample. Much faster than traditional scanning systems, the new resonance scanning systems are comparably as fast as spinning disk technology.
There are limits in terms of scan speed. “You don’t see people using 405 fps for high-speed events because most proteins introduced into cells for imaging tend to be green, yellow, red, and even some blue fluorophores, but they tend to be less stable and that’s a function of the protein itself, so they bleach faster,” Lopez explains.
Double-photon techniques are gaining in popularity but still do not exist on benchtop systems—they are years from commercialization. “Single-photon technology is more approachable, lasers are easier to work with; you’ll see benchtop instruments for doing routine confocal work,” Lopez says.
One most historically important secondary messenger to help peer into cells is calcium—concentration changes from inside to outside can be as high as 1,000x. “That’s a huge change,” Lopez says. During the infancy of LCI, calcium was the predominant imaging target. Today it is still an important assay, especially with the development of fluorescently labeled proteins that track calcium throughout the cell. Assays for other physiologically relevant metals have been commercialized, such as kits based on GCaMP.
Because of that, calcium is a very common secondary messenger in cell signaling. Calcium used to be the only thing that was imaged, and is still very important— many of the examples we use use calcium imaging. But now it’s just one of many things people look at. The proteins people have engineered over the past ten years are brighter, more stable, and more physiologically relevant (i.e., they don’t disturb cell machinery as much). People can do very high-speed, very bright, beautiful imaging because of the work through protein engineering. You can design an optical readout for pretty much any biological experiment you can think of. This has led to a great diversity of live-cell applications and experiments.
High-Resolution Electron Microscopy
Hitachi High Technologies America (Gaithersburg, MD) focuses primarily on electron microscopy (EM) and ion beam technology for high-resolution imaging that is beyond the capabilities of optical or confocal microscopy. The company has also advanced correlative light electron microscopy, which combines optical and EM methods to provide unique imaging perspectives.
Applications in LCI are trending toward what Phil Bryson, VP and general manager of the Nanotechnology Systems Division at Hitachi, calls a “correlation of techniques” where one technique alone is insufficient. “People want to go faster and see more detail. That’s where EM comes into play with cell imaging.”
EM is a black-and-white technique that provides excellent high-magnification resolution, whereas optical methods provide color and lower-magnification details. For example, during mitosis, optical microscopy can visualize spindle fibers during chromosome separation and EM can visualize individual centrioles or tubulin proteins.
Another interesting development involves the use of ionic liquids for sample preparation in scanning electron microscopy (SEM). Ionic liquids are salts that exist in the liquid state. Hitachi has introduced an organic liquid that is fluid at room temperature and, according to Bryson, as benign as table salt.
In LCI, ionic liquids allow imaging in a natural state with minimal sample prep. “One challenge with EM is imaging nonelectrically conductive objects,” Bryson explains. “When you hit it with electrons, the object charges up. Ionic liquids coating cells draw these charges away and permit imaging under vacuum.”
He describes cells as “balloons” filled with water that pop or shrink under the imaging conditions of EM. Ionic liquids maintain the cells’ structural integrity throughout the vacuuming process and without undo electron beam exposure to cells.
An additional benefit allows imaging under an optical system, followed by direct EM examination with minimal additional sample preparation.
A few weeks ago, Hitachi introduced an atmospheric scanning EM under the product name AeroSurf. The system behaves like a normal SEM when operating in vacuum mode, but can also image cells under less stressful atmospheric pressure.
Vacuum conditions are required for normal SEM operation due to molecular collision–induced electron scattering caused by air between the electron source and the sample. A silicon nitride membrane positioned just microns above the sample allows the electron source to remain under vacuum while the sample resides at atmospheric pressure. “The technique allows us to image samples in their natural state, without vacuum deformation artifacts,” Bryson says.
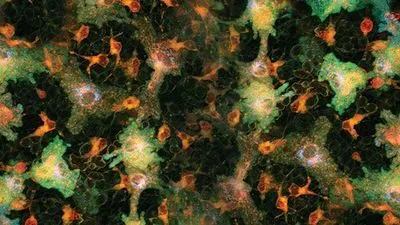
 Figure 1. Xenopus spp. tadpole epidermis acquired with 30x silicone oil objective NA 1.05 on an Olympus FluoView laser scanning confocal imaging system. Specimen labeled with methylene blue and malachite green. Scale bar indicates 100um.All images courtesy: James Lopez, Olympus Corporation of America Figure 1. Xenopus spp. tadpole epidermis acquired with 30x silicone oil objective NA 1.05 on an Olympus FluoView laser scanning confocal imaging system. Specimen labeled with methylene blue and malachite green. Scale bar indicates 100um.All images courtesy: James Lopez, Olympus Corporation of America |  Figure 2. Xenopus spp. tadpole epidermis acquired with 100x silicone oil objective NA 1.35 on an Olympus FluoView laser scanning confocal imaging system. Specimen labeled with methylene blue and malachite green. Scale bar indicates 30um. Figure 2. Xenopus spp. tadpole epidermis acquired with 100x silicone oil objective NA 1.35 on an Olympus FluoView laser scanning confocal imaging system. Specimen labeled with methylene blue and malachite green. Scale bar indicates 30um. |  Figure 3: Lemna minor leaf acquired with 30x silicone oil objective NA 1.05 on an Olympus FluoView laser scanning confocal imaging system. Scale bar indicates 100um. Figure 3: Lemna minor leaf acquired with 30x silicone oil objective NA 1.05 on an Olympus FluoView laser scanning confocal imaging system. Scale bar indicates 100um. |