 Perform XF analysis with an easy-to-use, reliable, validated, and supported cell count based normalization solution with the Agilent Seahorse XFe Analyzer and BioTek Cytation 1 Cell Imaging Multi-Mode Reader
Perform XF analysis with an easy-to-use, reliable, validated, and supported cell count based normalization solution with the Agilent Seahorse XFe Analyzer and BioTek Cytation 1 Cell Imaging Multi-Mode Reader
Problem: Normalization of functional biological data is a key component in the workflow for performing and/or subsequent analysis of raw data to ensure accurate and consistent interpretation of results. As a typical dataset usually contains more than one sample, and as researchers are almost always interested in making statistical comparisons between these samples, some form of normalization is usually required for most experiments performed. Whether comparing different cell types, genetic modifications, or compound treatments, the data must be normalized to a common shared parameter for correct comparison. Normalization of cellular assays can be applied on several levels, including cell number, genomic DNA, and total cellular protein.
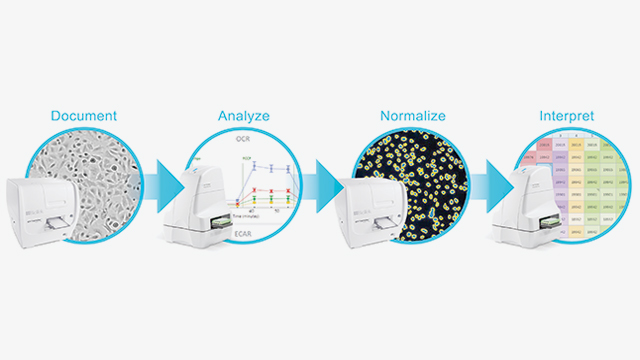
Solution: The solution described combines cellular metabolic analysis technology from Agilent and imaging technology from BioTek Instruments to create a standardized approach for comparing XF data sets, improving assay workflow, and applying normalization values to XF measurements. The novel integration of the Agilent Seahorse XFe Analyzer with the BioTek Cytation 1 Cell Imaging Multi-Mode Reader focuses on a method that uses cell number to normalize. The integrated system is optimized to automate and simplify the acquisition of brightfield (before and after XF assay) and fluorescence (after XF assay) images, via a unified controller that controls both the BioTek and Agilent Seahorse instruments. Specialized software then calculates the cell number in each microplate well, and seamlessly transfers the images and cell counts into the software for XF data normalization. The embedded brightfield images in the software provide visual feedback and quality control of cell seeding conditions, which improves live cell assay reproducibility and XF data quality. An “alert” within the software announces the availability of cell counts and images, and a heatmap display of cell counts enables easy evaluation of cell seeding consistency, making each assay run more robust. The incorporation of the high-quality imagery within the XF software adds another dimension to the data, as researchers can toggle between the XF data, brightfield images, and fluorescence images in a unified software experience. Referencing the images while analyzing XF data provides evidence and guidance on how to limit variability and improve the reproducibility of the assays. Applying a consistently generated cell count based normalization value ultimately makes interpreting the data and finding relationships amongst the data easier.
The benefits of this solution are:
- Simplified XF analysis with an easy-to-use, reliable, validated, and supported cell count based normalization solution
- Improved XF data interpretation by applying cell count numbers directly to your XF data enabling plate-plate, experiment-experiment, and well-well comparisons
- Documented cell culture condition throughout the XF assay to make better connections between cell seeding conditions and XF data, while providing guidance to detect statistical sources of errors
- Associated normalization values, brightfield and fluorescence images in the XF data analytical software package to provide evidence for making more meaningful biological conclusions
- Enhanced live cell assay reproducibility optimized for XF assays • Simplified normalization workflow with uncomplicated software and single controller operation to communicate data between both devices
For more information, please visit Agilent.com .












