Introduction
Liquid biopsies have emerged as a powerful and minimally invasive tool for detecting, classifying, and monitoring tumors, opening the door to early detection through widespread, routine multi-cancer screening. Detecting the low fraction of circulating tumor DNA (ctDNA) in liquid biopsy samples, however, requires higher sensitivity than the level obtained by traditional genetic mutation-based assays alone. Epigenetic markers, such as DNA methylation patterns, enable the capture of tissue- and state-specific signals. Multiomic analysis that incorporates both DNA variants and methylation data together can boost the sensitivity, accuracy, and confidence of ctDNA-based cancer analysis.
To realize the full potential of liquid biopsies, new workflows designed to support low inputs are needed to integrate highly sensitive detection methods, robust and reproducible laboratory procedures, and flexible, reliable data analysis. This article presents a streamlined, end-to-end workflow that combines genetic and epigenetic sequencing and analysis from a single library, providing a more comprehensive view of tumor biology. Library preparation and target enrichment for DNA variants and methylation markers are performed using the same DNA sample input, based on the novel Agilent Avida Duo technology—an innovative, targeted next-generation sequencing (NGS) approach offering high efficiency and sensitivity. Sequencing data analysis is optimized and streamlined with the SeqOne platform from FASTQ to report, which includes the SomaLBx pipeline for variant analysis and the SomaMethyl pipeline for methylation analysis. These integrated solutions transform complex multiomic data into relevant genomic and epigenomic insights.
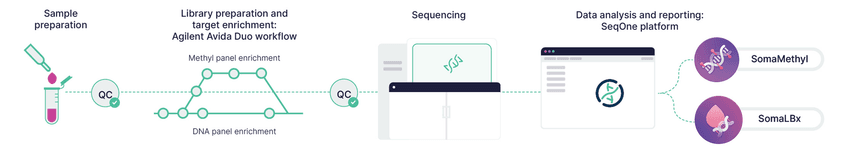
Figure 1. End-to-end workflow from liquid biopsy sample to data analysis.
CREDIT: Agilent
From one sample to two signals: method overview
The Avida Duo workflow takes a single sample, such as cell-free DNA (cfDNA), through library preparation and two enrichments—one targeting cancer-associated genes and the other targeting differentially methylated regions (DMRs). After sequencing the enriched libraries, the data are routed on the SeqOne platform into two analysis paths: a variant pipeline for markers such as SNVs, indels, CNAs, and fusions, and a methylation pipeline for CpG/DMR-level views and summary metrics. This keeps wet-lab handling lean and analysis purpose-built for each signal type.
Efficient target enrichment at the bench
The Avida hybridization chemistry has been optimized to handle the low amounts of input from liquid biopsy samples. Target enrichment is performed using an Avida pan-cancer catalog or a custom panel. The Avida Duo workflow can be completed in a single laboratory shift—typically eight to eight and a half hours—to produce enriched, sequencing-ready libraries (Figure 2).
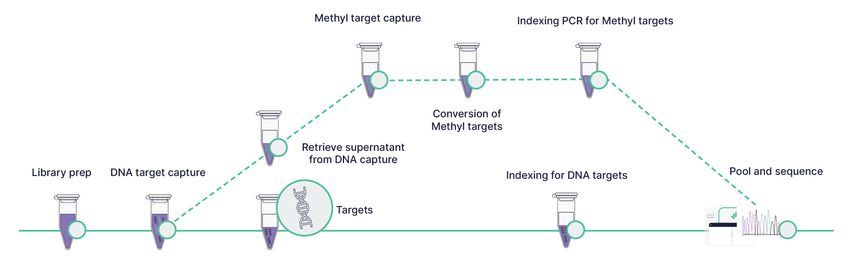
Figure 2. The Agilent Avida Duo workflow enables the detection of DNA variants and methylation markers from the same sample input, with a fast turnaround time.
CREDIT: Agilent
Reading methylation alongside variants
Adding methylation data improves tumor signal detection when variant allele fractions are low, dispersed, or confounded by competing signals. To enhance the precision of tumor-specific methylation pattern detection, the Avida platform introduces a single metric, the Avida Methylation Index (MI), that quantifies methylation status. The Avida MI provides a quantitative measure of methylation status across the DMRs targeted by the Agilent Avida Methyl 3400 DMR Cancer panel to support longitudinal tracking or group contrasts. These methylation data can be viewed in addition to variants to build confidence in calls or trends.
Advanced liquid biopsy and methylation analysis, and interpretation
Post-run, the data are split into two dedicated pipelines—one per signal type—so review can proceed in parallel.
Variant analysis. Using a pipeline optimized specifically for liquid biopsy samples processed with Avida DNA Cancer panels, SomaLBx delivers streamlined DNA sequencing data analysis integrated with the SeqOne platform. This solution enables molecular labs to:
• Efficiently process and analyze complex DNA sequencing data from cfDNA with optimized algorithms
• Accurately detect a wide range of genomic alterations, including SNVs, indels, CNAs, and fusions, critical for understanding tumor genomics
• Reduce turnaround times and enhance laboratory productivity with a rapid and user-friendly workflow
Methylation analysis. A second tailored pipeline, SomaMethyl, provides users with a streamlined analysis of targeted methylation sequencing data enriched by the Avida Methyl 3400 DMR Cancer panel. SomaMethyl outputs visualization of CpG island coverage, heatmaps of differential methylation, genome-wide profiles, and a comprehensive list of methylated regions with relevant QC metrics and the Avida MI value.
Conclusion
The sample-to-insight workflow described here provides streamlined capture at the bench to improve recovery and retention, parallel downstream analysis paths for variants and methylation to deliver consistent QC, cohort-ready views, and a concise methylation metric to support trends over time. Together, these elements form a practical, study-ready framework that teams can pilot quickly, tune with familiar QC thresholds, and scale on existing sequencing infrastructure. One input, two complementary signals, and a cohesive workflow built for real-world liquid biopsy research lay the groundwork for the data-rich evaluations and validations to come.
Use case examples
• Translational LBx method development, where variant-only assays underperform at very low variant allele frequency (VAF), and methylation can add orthogonal confidence
• Laboratories seeking to conserve input and unify review with a two-path analysis that lands in a single platform experience
Liquid biopsy and methylation workflow on SeqOne platform
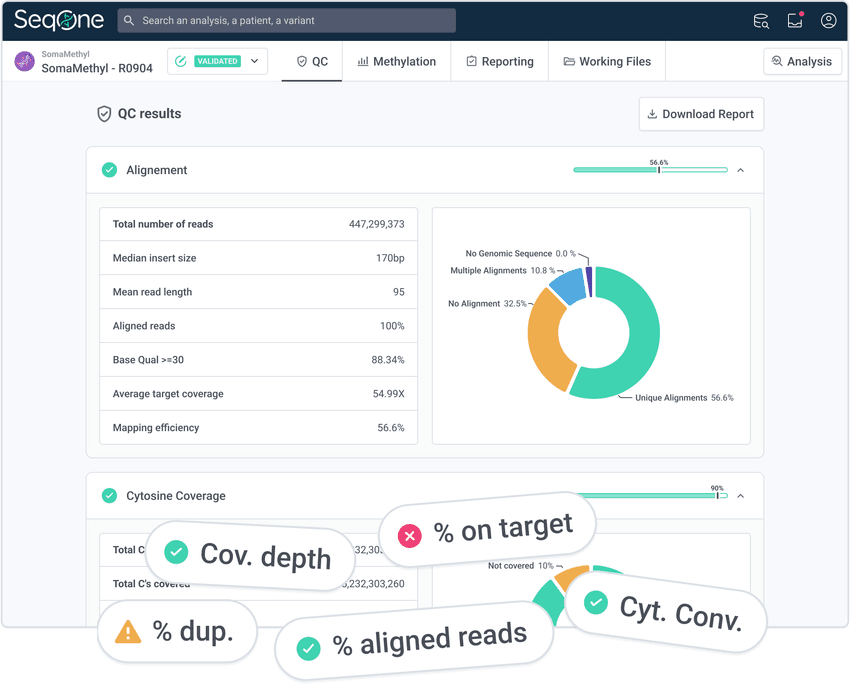
1. Initiate your analysis from FASTQ files from LBx or methylation samples
Leverage an optimized workflow for automatic alignment, quality control, and variant calling
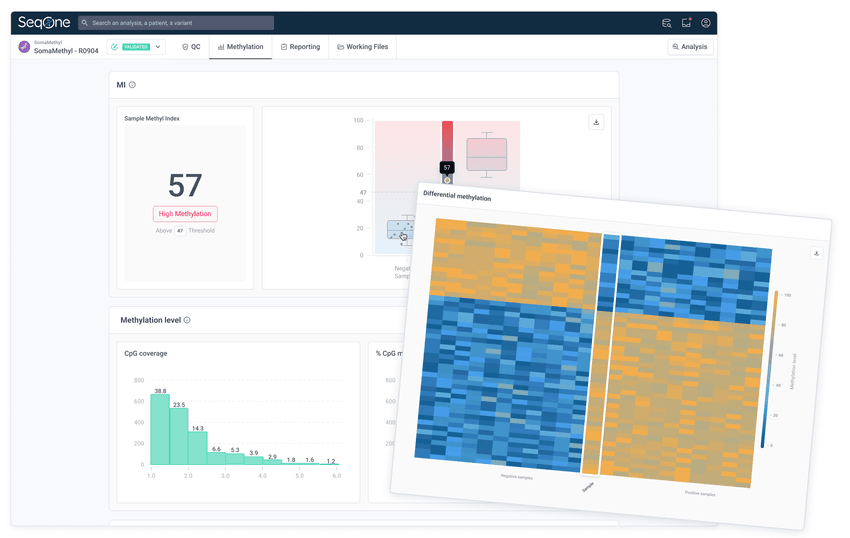
2. Analyze methylation
Get Avida Methylation Indexes and differential methylation patterns against cohorts of positive and negative samples
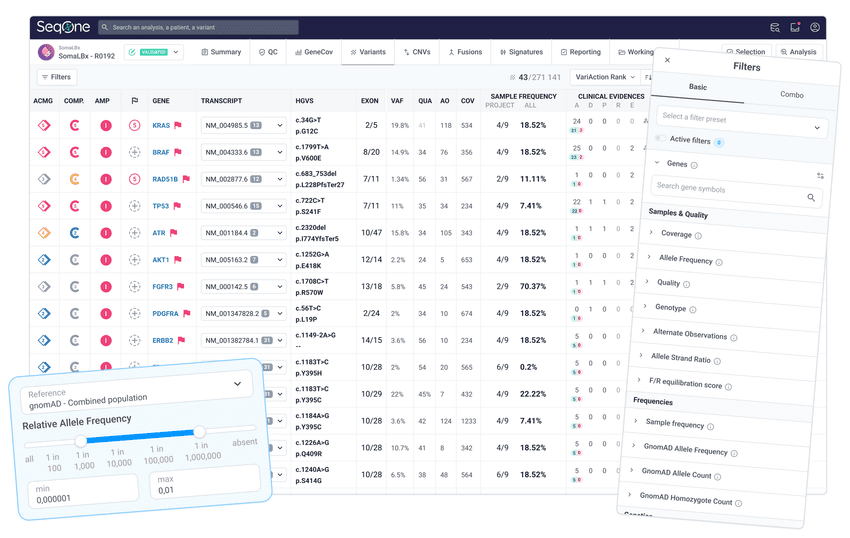
3. Filter and prioritize variants from LBx
Use prioritization algorithms and custom filters to reduce thousands of variants to a relevant set in one click
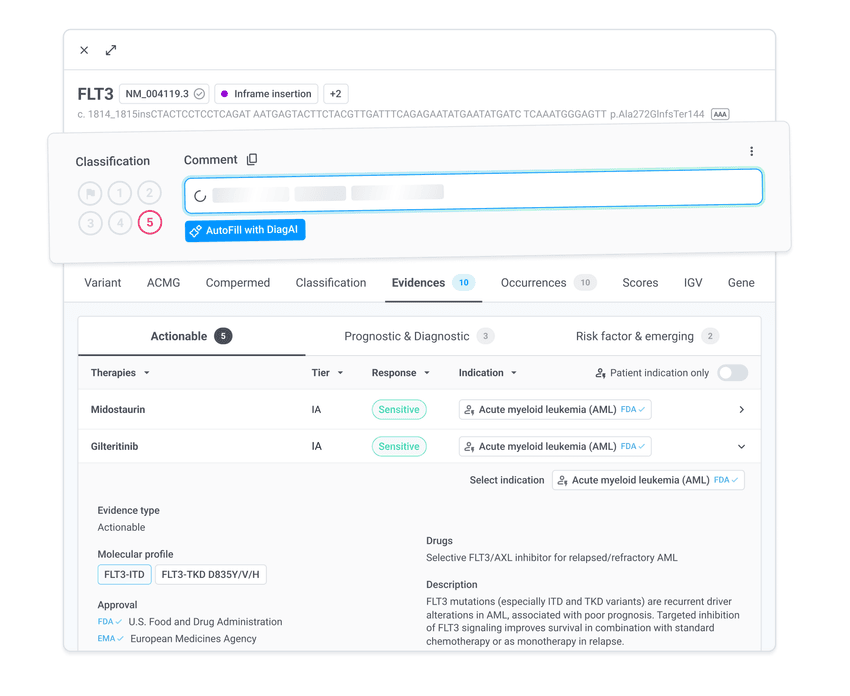
4. Curate variants
Leverage AI with the guideline-fed annotation drawer to classify, comment, and interpret your variants
Use the Variant Knowledge Base feature to track your in-house variant ranking
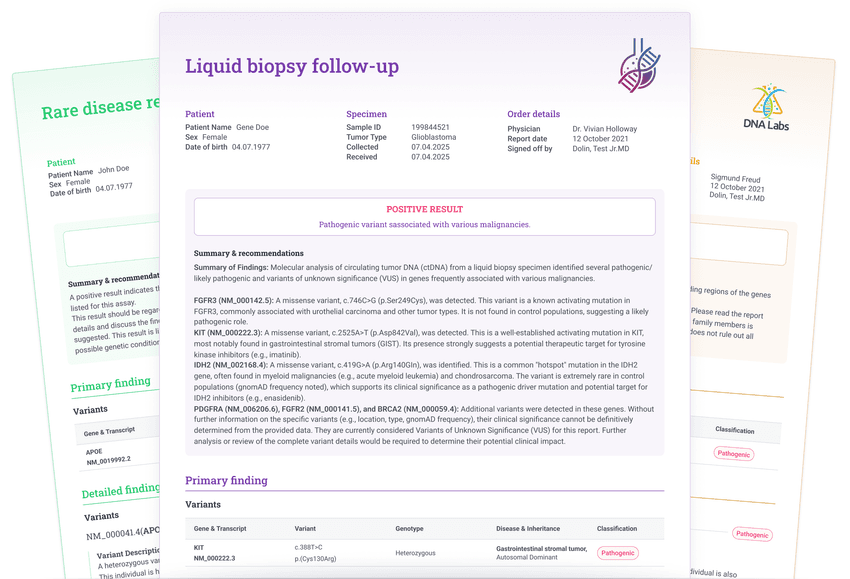
5. Generate reports
Automatically import all selected variants and sample metadata into comprehensive reports
For Research Use Only. Not for use in diagnostic procedures.