The integrity of scientific discovery hinges upon reproducibility, a core principle that dictates the consistency of experimental results. This consistency must hold when experiments are conducted by different personnel, in different settings, and using the same defined methods. Ensuring high reproducibility in academic research is paramount. It directly influences the credibility of published findings and the trajectory of translational science. For professionals operating within academic labs, the adoption of stringent quality assurance (QA) and quality control (QC) frameworks is not merely a best practice; it is an ethical and professional mandate. Failures in this area lead to resource waste, delays in medical and technological advancements, and a crisis of confidence in the scientific enterprise. Addressing the complexities of experimental variability requires a systematic, multi-faceted approach. This work transforms laboratory operations from ad-hoc processes into highly standardized systems.
Establishing comprehensive standard operating procedures and protocol management
Consistent outcomes in academic research begin with the standardization of every step, demanding detailed documentation that leaves no room for ambiguous interpretation.
A primary pillar of robust reproducibility is the establishment and scrupulous adherence to comprehensive standard operating procedures (SOPs). Within academic labs, SOPs must move beyond simple overviews. They must encompass granular detail on instrument calibration, reagent preparation, environmental controls, and execution specifics for every protocol. Documenting seemingly minor details can be the difference between a repeatable experiment and an irreproducible one. These details include incubation times (down to the second), buffer pH adjustments, or centrifugation temperature fluctuations.
The documentation system itself must be managed rigorously. Electronic lab notebooks (ELNs) are indispensable tools. They ensure version control and time-stamping, providing an audit trail for protocol modifications. Any change, no matter how small, must be logged, dated, and justified. Furthermore, SOPs should be reviewed and updated annually. This update should also occur whenever a major piece of equipment or methodology is introduced or retired. This process ensures the active protocol reflects current best practices within the academic lab.
Key documentation elements for reproducibility:
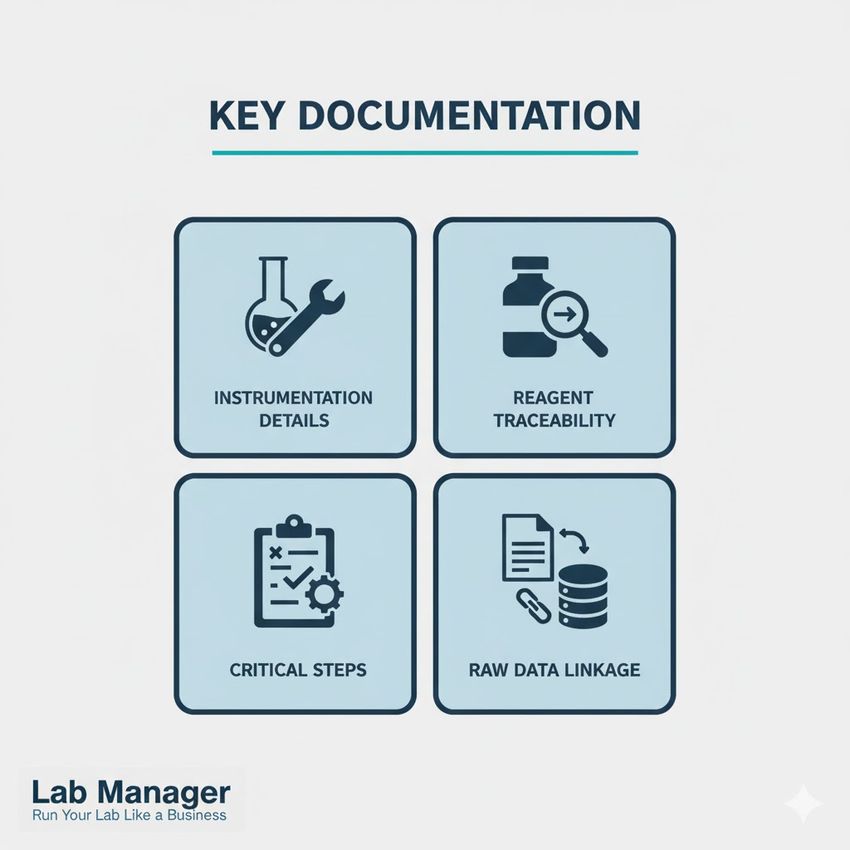
Mastering Documentation: The Four Pillars of Lab Quality. Ensure regulatory compliance and robust data integrity by focusing on these essential documentation checkpoints.
GEMINI (2025)
- Instrumentation details: Include manufacturer, model number, maintenance schedule, and last calibration date for all utilized equipment.
- Reagent traceability: Document lot numbers, expiry dates, supplier information, and storage conditions for all commercial and in-house reagents.
- Critical steps: Identify and highlight steps with high sensitivity to variation (e.g., cell counting, homogenization speed, mixing technique).
- Raw data linkage: Clearly link the executed protocol version to the corresponding raw data file generated from that specific experiment.
This detailed, structured approach aligns with recommendations from organizations like the National Institutes of Health (NIH), which stress rigor and reproducibility in grant-funded academic research.
Ensuring rigorous data stewardship and computational transparency
The reliability of results in academic research is intrinsically linked to the transparency and integrity of the data processing and analysis pipelines used.
In modern academic labs, experiments generate vast datasets, making data stewardship a critical component of reproducibility. Adopting the FAIR data principles—making data Findable, Accessible, Interoperable, and Reusable—provides a framework for handling large and complex datasets GO-FAIR Initiative. Data must be archived in designated, secure repositories with clear metadata tags detailing the experimental conditions and processing steps. This is a fundamental QA/QC measure.
Beyond storage, computational reproducibility is non-negotiable. All data analysis should be performed using documented, version-controlled scripts or code (e.g., R, Python, MATLAB). Computational environments like Jupyter notebooks or R Markdown are beneficial. They allow researchers to weave code, output, and narrative into a single document, making the entire analytical workflow transparent.
If data is cleaned or processed, the original raw data must be preserved and remain readily accessible. Processing choices, outlier removal, and statistical methods must be explicitly justified within the documentation. Failure to provide the original data and the exact code used for transformation and analysis is one of the most common breakdowns in reproducibility within academic research.
Data stage | Reproducibility requirement | Tools for implementation |
|---|---|---|
Collection | Automated logging of instrument output and metadata. | ELNs, Integrated instrument software. |
Storage | Secure, version-controlled archiving of raw and processed files. | Cloud storage, Institutional data repositories. |
Analysis | Public availability of all analysis code and software environment details. | Git/GitHub, Jupyter/R Markdown, Docker/Containerization. |
Reporting | Exact figures linked back to the originating data subsets. | Data visualization software with traceable sources. |
Implementing robust quality control for reagents and materials
Contamination and misidentification of biological and chemical materials present substantial threats to reproducibility, requiring proactive QA/QC procedures.
The quality of input materials is a frequent, yet often overlooked, variable affecting reproducibility in academic research. Academic labs must institute mandatory QA/QC checks for all critical reagents. For biological systems, this includes cell line authentication. This is a crucial step, given that the misidentification or contamination of cell lines by mycoplasma or other cells is a widespread issue. Authentication techniques like short tandem repeat (STR) profiling should be regularly employed, mirroring standards set by organizations like the American Type Culture Collection (ATCC).
Similarly, the specificity of antibodies—a cornerstone of numerous immunological and molecular assays—must be validated internally prior to use. Relying solely on manufacturer specifications is insufficient. Researchers should conduct western blots, immunoprecipitation, or knockout/knockdown experiments. This confirms target recognition under the specific experimental conditions employed.
For chemical reagents, batch variability must be documented. New lots of critical chemicals, enzymes, or media components should be tested alongside an established lot (the gold standard) in a relevant assay to verify equivalent performance. This verification must happen before they are fully implemented in academic research protocols. The consistent application of these QA/QC steps is foundational to minimizing unexpected variables.
Cultivating a culture of transparency and training for reproducibility
Achieving high reproducibility is ultimately a systemic and cultural challenge. It requires continuous professional development and open communication within academic labs.
Technical controls and documentation are effective only when supported by a culture that prioritizes rigor over speed. Academic labs must foster an environment where transparency and detailed reporting are rewarded. Furthermore, negative or unexpected results should be viewed as valuable data points, not failures to be ignored. Promoting open science practices, including pre-registration of studies, is becoming increasingly relevant for enhancing reproducibility in academic research.
Rigorous training programs are essential for new and existing personnel. Training must cover the technical skills needed for specific assays. It must also cover the philosophical importance of reproducibility and the practical application of the lab's QA/QC documentation. Technicians and researchers should be trained on the correct use of ELNs, version control systems, and data archiving protocols. Ongoing education ensures that the entire academic lab staff maintains proficiency in these critical areas. This commitment upholds the standard of quality essential for credible academic research.
Strengthening the scientific record through diligent QA/QC
The future progress of science and technology depends on the capacity for academic labs to consistently achieve high standards of reproducibility. The implementation of comprehensive SOPs, transparent data stewardship, and mandatory material QA/QC are not just compliance requirements but accelerators of scientific trust and discovery. The adoption of these stringent procedures moves the scientific community toward a more reliable and sustainable research model.
Frequently asked questions (FAQ)
What is the distinction between reproducibility and replicability in academic research?
Reproducibility refers to obtaining consistent results using the same materials, data, and code as the original study. Replicability refers to obtaining consistent results when a study is conducted again using different data, samples, or methods, but addressing the same scientific question. Both are vital to validating findings in academic research.
How do electronic lab notebooks (ELNs) improve data reproducibility and QA/QC?
ELNs fundamentally improve reproducibility by centralizing all experimental data, metadata, and protocols in a single, searchable platform. They automatically time-stamp and version-control documents, providing a clear, immutable audit trail that satisfies QA/QC requirements for all experiments conducted in academic labs.
Why is cell line authentication a required quality control measure for academic labs?
Cell line authentication is required because cell line cross-contamination (where one line is overgrown by another) is a pervasive problem that renders experimental results meaningless. Mandatory short tandem repeat (STR) profiling ensures the cell line used in academic research is precisely the one stated in the protocol, upholding the integrity of the data and improving reproducibility.
This article was created with the assistance of Generative AI and has undergone editorial review before publishing.