Problem:
It is difficult to use traditional, one-color immunohistochemistry to identify multiple cell types and targets in tumors due to the paucity of biopsy tissue and its heterogeneous nature.
Oncology, fibrosis, respiratory, and autoimmune disease are complex conditions involving multiple cell types and populations. Identifying the cell types present, or the therapeutic target of interest, in diseased tissues is routinely accomplished with single color immunohistochemistry (IHC). This approach can determine the location and, to a limited extent, the prevalence of a known cell type or therapeutic target. It can also identify target-expressing tumor cells or immune cells in a tissue sample based on review of the staining pattern relative to tissue morphology. However, identification of multiple cell types and targets using this single color approach requires assays to be run using large numbers of sequential tissue sections. In a clinical environment, often only small and very precious biopsy samples are available, from which it may be difficult to get sufficient suitable sections. Given the heterogeneous nature of a tumor, this approach may not lend itself to accurate overlay of the resulting images to allow assessment of marker co-localization at the cellular level.
Solution:
Multiple markers can be immunostained in a single tissue section using multiplexed IHC (mIHC). Traditional chromogenic mIHC relies on each marker being identified using antibodies raised in different animal species or different isotypes and enzymatic deposition of chromogen on the tissue section. This limits the number of markers or targets that can be detected in a single slide. Deconvolution of chromogenic immunostaining is also typically limited to four colors due to the relatively narrow visible spectrum.
Converting the IHC detection system to tyramide signal amplification, which covalently labels the tissue section with fluorescent immunostains for each marker, enables the process to be serially repeated through several rounds of antibody stripping. It also allows multiple antibodies from a single species to be used. In addition, the fluorescence spectrum permits more colors to be introduced.
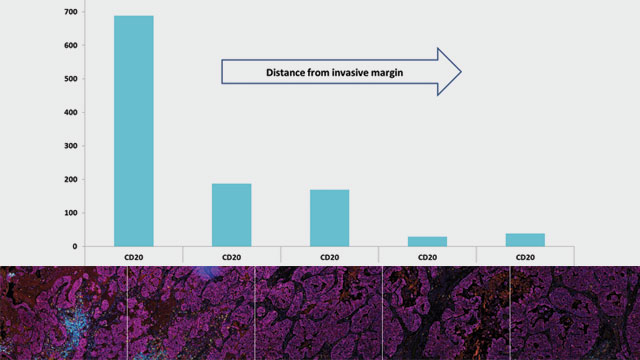
Fluorogenic mIHC enables up to six markers to be immunostained in a single tissue section. This level of multiplexing permits high-content data to be generated from one tissue section, effectively reducing the amount of tissue required. Multiple cell populations and targets can then be identified, and each target can be localized to individual or even multiple cell populations using digital image analysis and pathology, allowing deeper mining of the multiple immunostains to identify inter-cell spatial relationships. Resulting data can be further interrogated to determine if those relationships change by locale within the sample (e.g. tumor versus stroma). One point to note, assays requiring multiple rounds of antibody binding, detection, and stripping have to be systematically developed to ensure robust and optimal detection of every marker in the panel.
Generating high-content data to identify the presence and juxtaposition of key cell types and targets in complex diseases enables the interrogation of pre- and post-treatment biopsy samples from responding and non-responding patients. This can provide information on key features / biologic profiles that will predict whether a therapeutic is effective in a particular disease and patient population.
Multiplexed IHC, combined with sophisticated digital analysis, is a powerful approach for interrogating the biology of complex disease, especially when there is limited clinical material available.
Visit www.bioivt.com to learn more.











