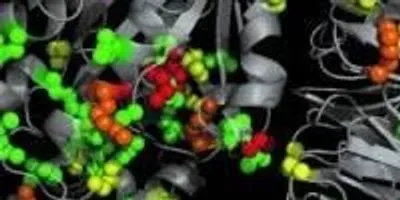
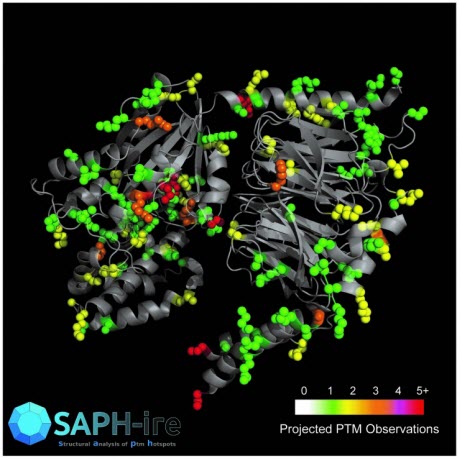
 SAPH-ire analyzes existing data repositories of protein modifications and 3D protein structures to help scientists identify and target research on “hotspots” likely to be important for biological function. Shown is the structural projection of PTM hotspots onto the 3D structure of a heterotrimeric G protein. Regions of the protein that are modified are color coded to indicate observation frequency. SAPH-ire integrates these and other features to predict which modifications are most likely to impact the function of the protein.Photo Credit: Torres LaboratoryKnown as SAPH-ire (Structural Analysis of PTM Hotspots), the tool could accelerate the search for potential new drug targets on protein structures, and lead to a better understanding of how proteins communicate with one another inside cells. SAPH-ire has been tested on a well studied class of proteins involved in cellular communication, where it correctly predicted a previously-unknown regulatory element.
SAPH-ire analyzes existing data repositories of protein modifications and 3D protein structures to help scientists identify and target research on “hotspots” likely to be important for biological function. Shown is the structural projection of PTM hotspots onto the 3D structure of a heterotrimeric G protein. Regions of the protein that are modified are color coded to indicate observation frequency. SAPH-ire integrates these and other features to predict which modifications are most likely to impact the function of the protein.Photo Credit: Torres LaboratoryKnown as SAPH-ire (Structural Analysis of PTM Hotspots), the tool could accelerate the search for potential new drug targets on protein structures, and lead to a better understanding of how proteins communicate with one another inside cells. SAPH-ire has been tested on a well studied class of proteins involved in cellular communication, where it correctly predicted a previously-unknown regulatory element.
“SAPH-ire predicts positions on proteins that are likely to be important for biological function based on how many times those parts of the proteins have been found in a chemically-modified state when they are taken out of a cell,” explained Matthew Torres, an assistant professor in the School of Biology at the Georgia Institute of Technology. “SAPH-ire is a tool for discovery, and we think it will lead to a new understanding of how proteins are connected in cells.”
The tool and its proof-of-concept testing were reported June 12 in the journal Molecular and Cellular Proteomics. The research was supported by the National Institutes of Health’s National Institute of General Medical Sciences (NIGMS) and Georgia Tech.
Through modern mass spectrometry proteomics techniques, scientists have identified more than 300,000 post-translational modifications (PTMs) in different families of proteins across numerous species. These PTMs come in many forms, resulting from the action of different enzymes, and are often indicators of how and where proteins contact one another to bring about different cell behaviors. The number of PTMs detected by mass spectrometry has grown so rapidly that researchers experimentally investigating the function of the modifications have been unable to keep up.
“Mass spectrometry is so effective that it has created an exponential curve in the knowledge of how proteins are modified,” said Torres. “The rate at which we can detect new PTMs has now far surpassed the rate at which we can understand what they do, from a classical biochemical approach. You have so much information that you don’t know where to begin.”
But that’s exactly where SAPH-ire begins. Aimed at bridging the gap between PTM detection and analysis of function, SAPH-ire collects non-redundant and experimentally verified PTM data across all known members of a protein family. Since members of a protein family share the same or similar protein structures, PTMs found within the family can be related to one another in three-dimensional space to produce a set of observed PTM frequencies, termed “hotspots.”
 Georgia Tech researchers (l-r) Shilpa Choudhury, Henry Dewhurst and Matthew Torres show a diagram of how their SAPH-ire tool works. SAPH-ire analyzes existing data repositories of protein modifications and 3D protein structures to help scientists identify and target research on “hotspots” likely to be important for biological function.Photo Credit: John Toon, Georgia TechThe PTM hotspots are projected onto 3D protein structures available in the Protein Data Bank (PDB), which allows the entire set of family-specific PTMs to be visualized on any protein structure that is representative for the family. Once projected there, SAPH-ire integrates multiple quantitative features from each hotspot to create a PTM “Functional Potential Score.” Each PTM hotspot can then be ranked in order of highest to lowest potential for having significant biological function.
Georgia Tech researchers (l-r) Shilpa Choudhury, Henry Dewhurst and Matthew Torres show a diagram of how their SAPH-ire tool works. SAPH-ire analyzes existing data repositories of protein modifications and 3D protein structures to help scientists identify and target research on “hotspots” likely to be important for biological function.Photo Credit: John Toon, Georgia TechThe PTM hotspots are projected onto 3D protein structures available in the Protein Data Bank (PDB), which allows the entire set of family-specific PTMs to be visualized on any protein structure that is representative for the family. Once projected there, SAPH-ire integrates multiple quantitative features from each hotspot to create a PTM “Functional Potential Score.” Each PTM hotspot can then be ranked in order of highest to lowest potential for having significant biological function.
“We have gone through all of what might be considered the meta-data that exists in the public domain, collected all the PTMs and all the structures, then organized them into their specific protein families,” Torres explained. “We are looking at PTMs through time, in a sense, because we have information from organisms that are evolutionarily distant from each other, though their proteins are related as members of a protein family.”
To prioritize research with the most significant potential impact, scientists might examine PTM hotspots that SAPH-ire identifies as having high function potential, but no known function.
Torres’ lab has been investigating unique families of “G” proteins, some of which cooperate with cell surface receptors that control the binding of hormones and neurotransmitters, as well as a majority of pharmaceutical drugs. Because of their importance to therapeutics, these proteins have been extensively studied over a period of 50 years or so. Using SAPH-ire, the researchers discovered something surprising about this group of protein families.
“We discovered a new regulatory element within a specific G protein family that has been largely ignored because it’s pretty unimpressive from a purely structural viewpoint,” Torres said. “SAPH-ire predicted that this element was going to be important from a modification point of view, and we confirmed experimentally that it was.”
SAPH-ire was conceived by Torres and developed by him and graduate student Henry Dewhurst, while experimental validation of the tool was accomplished by graduate student Shilpa Choudhury. Their next step is to develop collaborations with scientists who will try it out on the protein families they study. The Georgia Tech researchers are also creating a database that other protein scientists can query to help them identify and prioritize PTM hotspots, and they expect to see their program become part of informatics systems used to analyze large volumes of proteomics data emerging from labs around the world.
“SAPH-ire will help bring meaning and context to all the data that is being produced about PTMs,” Torres said. “Connecting SAPH-ire to other programs that convert mass spec data into actual PTM data could provide immediate biological relevance and prioritization for biochemists and others. It is likely to expose many new and unsuspected relationships between protein modification, protein structure and function.”
This research was supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS), under grant number 5R00 GM094533-05. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CITATION: Henry M. Dewhurst, Shilpa Choudhury and Matthew P. Torres, “Structural Analysis of PTM Hotspots (SAPH-ire) – a Quantitative Informatics Method Enabling the Discovery of Novel Regulatory Elements in Protein Families,” (Molecular and Cellular Proteomics, 2015). http://dx.doi.org/10.1074/mcp.M115.051177