Laboratory professionals increasingly rely on high-resolution analytical technologies to decipher the complex molecular interactions underlying disease states and therapeutic interventions. Mass spectrometers serve as the cornerstone of this investigative process, offering the distinct ability to identify and quantify biomolecules with unparalleled precision.
By enabling deep insights into the proteome and the metabolome, these instruments allow researchers to move beyond simple genetic screening and understand the functional output of cellular systems. The integration of mass spectrometers into early-stage drug discovery pipelines facilitates the rapid identification of novel drug targets and the validation of biomarkers, significantly reducing the time and cost associated with bringing new therapeutics to market.
Proteomics and Mass Spectrometers for Target Identification
High-resolution mass spectrometry provides the necessary depth to characterize protein expression, modification, and interaction networks within biological samples.
Proteomics, the large-scale study of proteins, has evolved from 2D gel electrophoresis to sophisticated liquid chromatography-mass spectrometry (LC-MS) workflows. In the context of drug discovery, mass spectrometers allow for the comprehensive analysis of the "druggable" proteome. This process typically involves bottom-up approaches where proteins are enzymatically digested into peptides before analysis.
The resulting spectral data facilitates the sequencing of peptides and the subsequent identification of the parent proteins. This capability is critical for target deconvolution—identifying the molecular mechanism behind phenotypic hits where a compound shows therapeutic activity but its target remains unknown.
Furthermore, post-translational modifications (PTMs) such as phosphorylation, glycosylation, and ubiquitination dictate protein function and localization. Aberrant PTMs often drive disease pathology, particularly in oncology and neurodegeneration. Modern mass spectrometers possess the resolution required to detect these subtle mass shifts, enabling researchers to map PTM sites precisely. For instance, monitoring changes in phosphorylation states before and after drug treatment helps delineate signaling pathways affected by a kinase inhibitor.
The Human Proteome Organization (HUPO) highlights the importance of standardized MS protocols in ensuring reproducibility across these complex datasets. Recent guidelines from the HUPO Human Proteome Project (HPP), published in Nature Communications, emphasize stringent data quality metrics to reduce false discovery rates in protein identification.
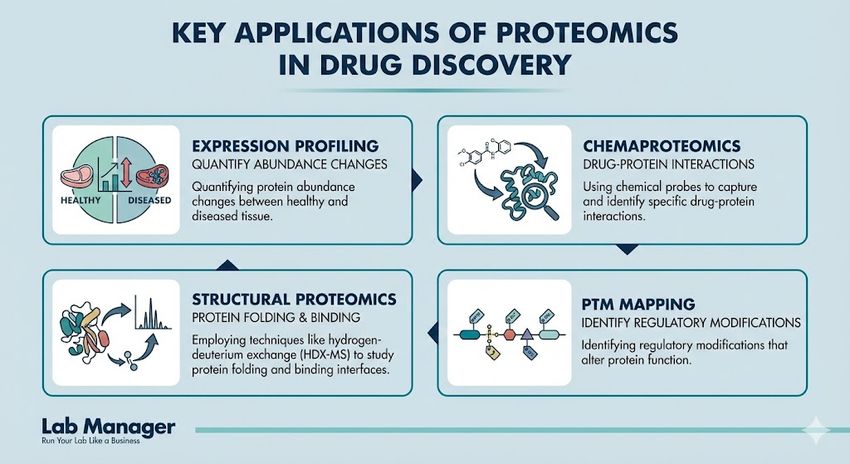
Key applications of proteomics in drug discovery include:
Proteomics is no longer just a buzzword—it is a critical engine driving modern drug discovery.
GEMINI (2026)
- Expression profiling: Quantifying protein abundance changes between healthy and diseased tissue.
- Chemaproteomics: Using chemical probes to capture and identify specific drug-protein interactions.
- Structural proteomics: Employing techniques like hydrogen-deuterium exchange (HDX-MS) to study protein folding and binding interfaces.
- PTM mapping: Identifying regulatory modifications that alter protein function.
Metabolomics Profiling with Mass Spectrometers for Biomarker Discovery
Metabolic phenotyping via mass spectrometry offers a real-time snapshot of cellular physiology and drug response.
While proteomics looks at the functional machinery of the cell, metabolomics examines the ultimate output—small molecule metabolites. Mass spectrometers are uniquely suited for this task due to their ability to detect diverse chemical classes, from lipids and amino acids to sugars and organic acids. In drug discovery, metabolomics serves two primary functions: efficacy monitoring and toxicity screening. By comparing the metabolic footprint of biofluids (plasma, urine) or tissues from treated versus untreated subjects, laboratory professionals can identify biomarkers that predict therapeutic response.
Targeted metabolomics focuses on a defined set of known metabolites, offering high sensitivity and quantitative accuracy. This approach is often used to validate specific biomarkers during clinical trials according to FDA Bioanalytical Method Validation guidelines. Conversely, untargeted metabolomics provides a global survey of the metabolome, useful for hypothesis generation and discovering unexpected off-target effects. For example, a rise in specific liver enzymes or bile acids detected via MS can signal potential hepatotoxicity early in the development process, long before clinical symptoms manifest.
Regulatory bodies increasingly encourage the use of genomic and metabolic biomarkers to guide drug development. The FDA’s Biomarker Qualification Program and ICH E16 guidelines provide specific frameworks for qualifying these biomarkers for regulatory decision-making, emphasizing the need for robust analytical validation.
Advantages of MS-based metabolomics include:
- High sensitivity: Detection of low-abundance metabolites in complex biological matrices.
- Wide dynamic range: Simultaneous analysis of major metabolites and trace signaling molecules.
- Speed: Rapid sample throughput suitable for screening large compound libraries.
- Versatility: Applicability to various sample types, including cell culture media, biofluids, and solid tissues.
Technological Advancements in Mass Spectrometers for Drug Discovery
Recent innovations in ionization sources and mass analyzers have dramatically expanded the analytical boundaries for laboratory professionals.
The utility of mass spectrometers in drug discovery directly correlates with advancements in instrument hardware. The shift from low-resolution instruments to high-resolution accurate mass (HRAM) systems, such as Orbitrap and Q-TOF (Quadrupole Time-of-Flight) analyzers, has revolutionized data quality. These systems offer resolving power sufficient to distinguish between isobaric compounds—molecules with the same nominal mass but different chemical formulas. This capability is essential in metabolomics, where distinguishing a drug metabolite from an endogenous molecule requires extreme precision.
Ion mobility spectrometry (IMS) represents another leap forward, adding a dimension of separation based on the shape and size of gas-phase ions. IMS allows researchers to calculate Collisional Cross-Section (CCS) values, a standardized parameter that is independent of chromatographic conditions. This adds a critical layer of confidence to compound identification, helping to separate isomers and reduce background noise, significantly enhancing peak capacity in complex proteomic digests.
Additionally, advancements in ionization techniques, such as Desorption Electrospray Ionization (DESI), allow for mass spectrometry imaging (MSI). MSI enables researchers to visualize the spatial distribution of drugs and metabolites directly within tissue sections, providing critical data on drug distribution and accumulation in target organs without the need for radioactive labeling.
Operational considerations for modern MS labs involve:
- Maintenance schedules: Regular calibration and cleaning to maintain mass accuracy.
- Source selection: Choosing between Electrospray Ionization (ESI) for polar compounds and Atmospheric Pressure Chemical Ionization (APCI) for non-polar analytes.
- Coupling technologies: Integrating MS with Ultra-High-Performance Liquid Chromatography (UHPLC) or Capillary Electrophoresis (CE) for optimal separation.
Integrating Mass Spectrometry Data for Multi-Omics Systems Biology
Combining proteomic and metabolomic datasets provides a holistic view of biological systems that accelerates decision-making.
Data generated by mass spectrometers in isolation offers limited visibility; the true power lies in integration. Systems biology approaches combine proteomic and metabolomic data to construct detailed models of disease networks. This multi-omics strategy allows researchers to trace the flow of information from protein expression to metabolic output. For example, identifying an upregulated enzyme via proteomics while simultaneously observing an increase in its catalytic product via metabolomics provides strong evidence for pathway activation.
This integrated approach helps mitigate false positives inherent in single-omics studies. If a proteomic change does not result in a corresponding metabolic phenotypic shift, the target may not be biologically relevant for the disease phenotype. Conversely, metabolic changes can point upstream to regulatory proteins that might have been missed in the initial proteomic screen.
The implementation of such comprehensive workflows requires sophisticated bioinformatics pipelines capable of aligning disparate datasets. Guidelines from industry consortia emphasize the importance of data standardization formats (such as mzML) to facilitate this cross-domain analysis.
Ensuring Regulatory Compliance for Mass Spectrometers in the Lab
Strict adherence to data integrity standards is essential when utilizing mass spectrometers for regulated drug development activities.
As mass spectrometers move from exploratory research into regulated environments like GLP (Good Laboratory Practice) toxicology and GMP (Good Manufacturing Practice) quality control, compliance becomes paramount. Laboratory professionals must ensure that all software controlling these instruments adheres to regulations such as FDA 21 CFR Part 11 regarding electronic records and signatures. This involves maintaining audit trails that log every user action, from method modification to data reprocessing.
Ensuring data integrity requires alignment with ALCOA+ principles (Attributable, Legible, Contemporaneous, Original, Accurate), a standard framework utilized during regulatory inspections. Validation of bioanalytical methods—proving that the instrument measures what it claims to measure with specific accuracy and precision—is a critical requirement outlined by the FDA and the OECD Principles of Good Laboratory Practice. Ensuring that mass spectrometers are properly qualified (IQ/OQ/PQ) and that system suitability tests are performed daily safeguards the reliability of the data used to make critical safety and efficacy decisions.
Conclusion: The Future of Mass Spectrometers in Drug Discovery
The continued evolution of mass spectrometers ensures their place as central components in the drug discovery ecosystem. By providing deep, quantitative insights into proteomics and metabolomics, these instruments allow laboratory professionals to unravel complex disease mechanisms and identify promising therapeutic targets with greater confidence. As hardware sensitivity improves and bioinformatics tools become more sophisticated, the integration of MS-based multi-omics will become standard practice, ultimately leading to safer, more effective drugs reaching the patient.
This article was created with the assistance of Generative AI and has undergone editorial review before publishing.















