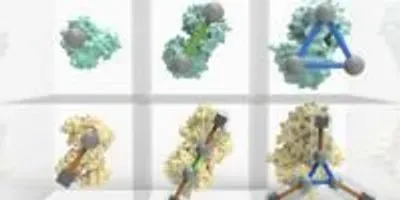
Image courtesy of the University of CambridgeA new ‘periodic table’ of protein complexes, devised by an interdisciplinary team of researchers, provides a unified way to classify and visualize protein complexes, which drive a huge range of biological processes, from DNA replication to catalyzing metabolic reactions.
Image courtesy of the University of CambridgeA new ‘periodic table’ of protein complexes, devised by an interdisciplinary team of researchers, provides a unified way to classify and visualize protein complexes, which drive a huge range of biological processes, from DNA replication to catalyzing metabolic reactions.
The table, published in the journal Science, offers a new way of looking at almost all known molecular structures and predicting how new ones could be made, providing a valuable tool for research into evolution and protein engineering.
By using the table, researchers are able predict the likely forms of protein complexes with unknown structure, estimate the feasibility of entirely new structures, and identify possible errors in existing structural databases. It was created by an interdisciplinary team led by researchers at the University of Cambridge and the Wellcome Genome Campus.
Almost every biological process depends on proteins interacting and assembling into complexes in a specific way, and many diseases, such as Alzheimer’s and Parkinson’s, are associated with problems in complex assembly. The principles underpinning this organization are not yet fully understood, but the new periodic table presents a systematic, ordered view on protein assembly, providing a visual tool for understanding biological function.
“We’re bringing a lot of order into the messy world of protein complexes,” said the paper’s lead author Sebastian Ahnert of Cambridge’s Cavendish Laboratory, a physicist who regularly tangles with biological problems. “Proteins can keep combining in these simple ways, adding more and more levels of complexity and resulting in a huge variety of structures. What we’ve made is a classification based on underlying principles that helps people get a handle on the complexity.”
The exceptions to the rule are interesting in their own right, added Ahnert, and are the subject of continuing studies.
“Evolution has given rise to a huge variety of protein complexes, and it can seem a bit chaotic,” said study co-author Joe Marsh, formerly of the Wellcome Genome Campus and now of the MRC Human Genetics Unit at the University of Edinburgh. “But if you break down the steps proteins take to become complexes, there are some basic rules that can explain almost all of the assemblies people have observed so far.”
Ballroom dancing can be seen as an endless combination of riffs on the waltz, fox trot and cha-cha. Similarly, the ‘dance’ of protein complex assembly can be seen as endless variations on dimerization (one doubles, and becomes two), cyclization (one forms a ring of three or more) and subunit addition (two different proteins bind to each other). Because these happen in a fairly predictable way, it’s not as hard as you might think to predict how a novel protein would form.
Some protein complexes, called homomers, feature multiple copies of a single protein, while others, called heteromers, are made from several different types of proteins. The table shows that there is a very close relationship between the possible structures of heteromers and homomers. In fact, the vast majority of heteromers can be thought of as homomers in which the single protein is replaced by a repeated unit of several proteins. The table was constructed using computational analysis of a large database of protein-protein interfaces.
“By analyzing the tens of thousands of protein complexes for which three-dimensional structures have already been experimentally determined, we could see repeating patterns in the assembly transitions that occur–and with new data from mass spectrometry we could start to see the bigger picture,” said Walsh.
“The core work for this study is in theoretical physics and computational biology, but it couldn’t have been done without the mass spectrometry work by our colleagues at Oxford University,” said Sarah Teichmann, Research Group Leader at the European Bioinformatics Institute (EMBL-EBI) and the Wellcome Trust Sanger Institute. “This is yet another excellent example of how extremely valuable interdisciplinary research can be.”
A new ‘periodic table’ of protein complexes, devised by an interdisciplinary team of researchers, provides a unified way to classify and visualize protein complexes, which drive a huge range of biological processes, from DNA replication to catalyzing metabolic reactions.
The table, published in the journal Science, offers a new way of looking at almost all known molecular structures and predicting how new ones could be made, providing a valuable tool for research into evolution and protein engineering.
By using the table, researchers are able predict the likely forms of protein complexes with unknown structure, estimate the feasibility of entirely new structures, and identify possible errors in existing structural databases. It was created by an interdisciplinary team led by researchers at the University of Cambridge and the Wellcome Genome Campus.
To continue reading this article, sign up for FREE to

Membership is FREE and provides you with instant access to eNewsletters, digital publications, article archives, and more.