Health Science

Study: Boosting Gut Bacteria Defense System may Lead to Better Treatments for Bloodstream Infections
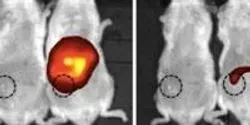
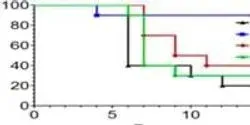
An upset in the body’s natural balance of gut bacteria that may lead to life-threatening bloodstream infections can be reversed by enhancing a specific immune defense response, UT Southwestern Medical Center researchers have found.

In a stunning discovery that overturns decades of textbook teaching, researchers at the University of Virginia School of Medicine have determined that the brain is directly connected to the immune system by vessels previously thought not to exist.

Grass plants can bind, uptake and transport infectious prions, according to researchers at the University of Texas Health Science Center at Houston (UTHealth). The research was published online in the latest issue of Cell Reports.

When people become severely burned, there are limited options available to repair the skin and tissue that have been affected. People must undergo painful and tedious surgery and skin transplants to repair the damaged skin and tissues. Stratatech Corp., a Madison, WI based company, has been focused on creating new treatment options for severe burns and recently developed new human skin tissue called StrataGraft that was used to heal severe burn wounds in 30 patients during a recent clinical trial.